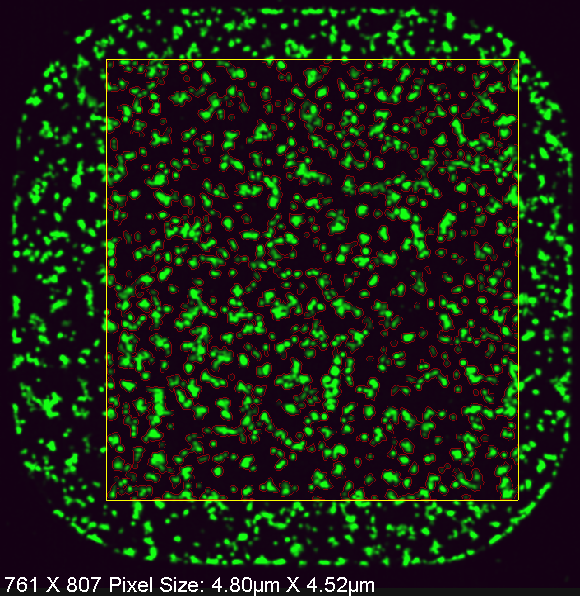
 Example of segmentation |
Cell-based screens are powerful because a “wider net” can be casted; i.e., an entire pathway can be interrogated in a single drug or siRNA screen. In addition, the identified drug hits have already passed a major hurdle which typically limits drug development: drug solubility and ability to penetrate cell membranes. This is because these are almost always pre-requisites for appearing as a hit in a phenotype-based cellular screen, since they drugs typically are added directly to the cell culture medium. However, cell-based screens can be quite difficult to design, miniaturize and optimize, in contrast to in vitro screens. This is largely because the “unknowns” of cancer cells must be factored in, and also because sophisticated imaging techniques are needed for the phenotypic read-out. |
 automated clone ID |
Our laboratory has developed a number of unique tools for this purpose. For examples, we have developed protocols for the automated imaging and detection of IR-induced DNA repair protein foci formation by confocal microscopy in 384-well plates (photo 1). We have also developed tools to mark live cells with nuclear localized, ligand-inducible fluorescent proteins (photo 2). In the latter case, we can use this technology to detect live single cell clones growing in microplates (as in photo 2). These are two examples, of many, that we in developed in our laboratory. We welcome new approaches and ideas in this project focus area. |


